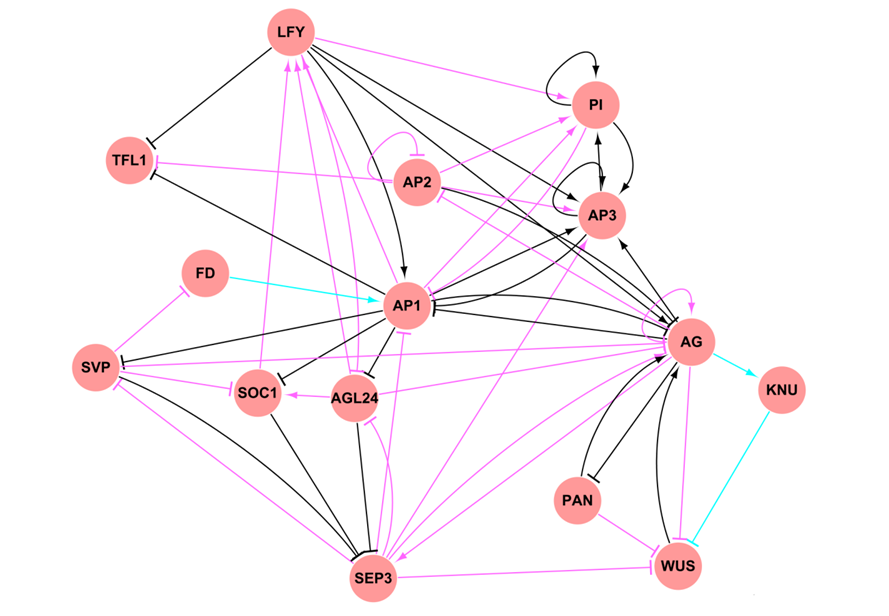
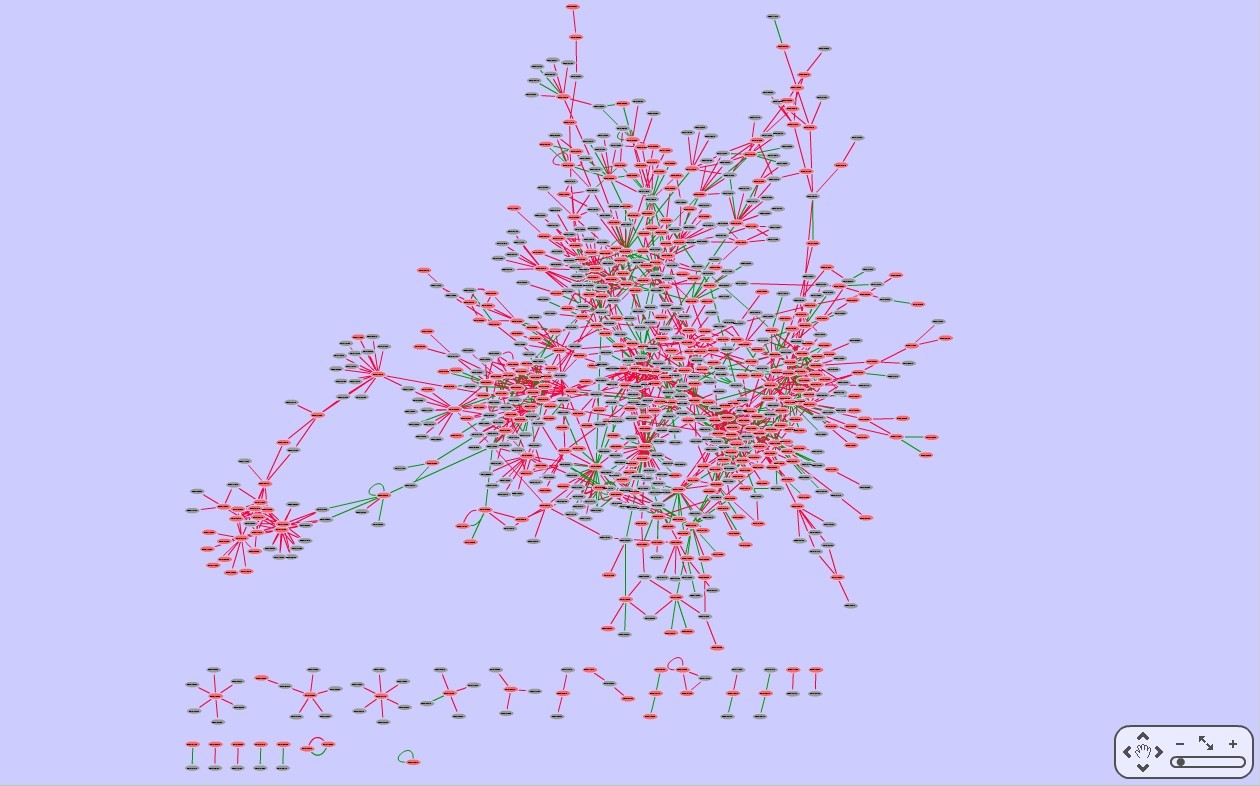
- Based on a systematic literature mining, we curate a high-confidence Arabidopsis transcriptional regulatory map (download).
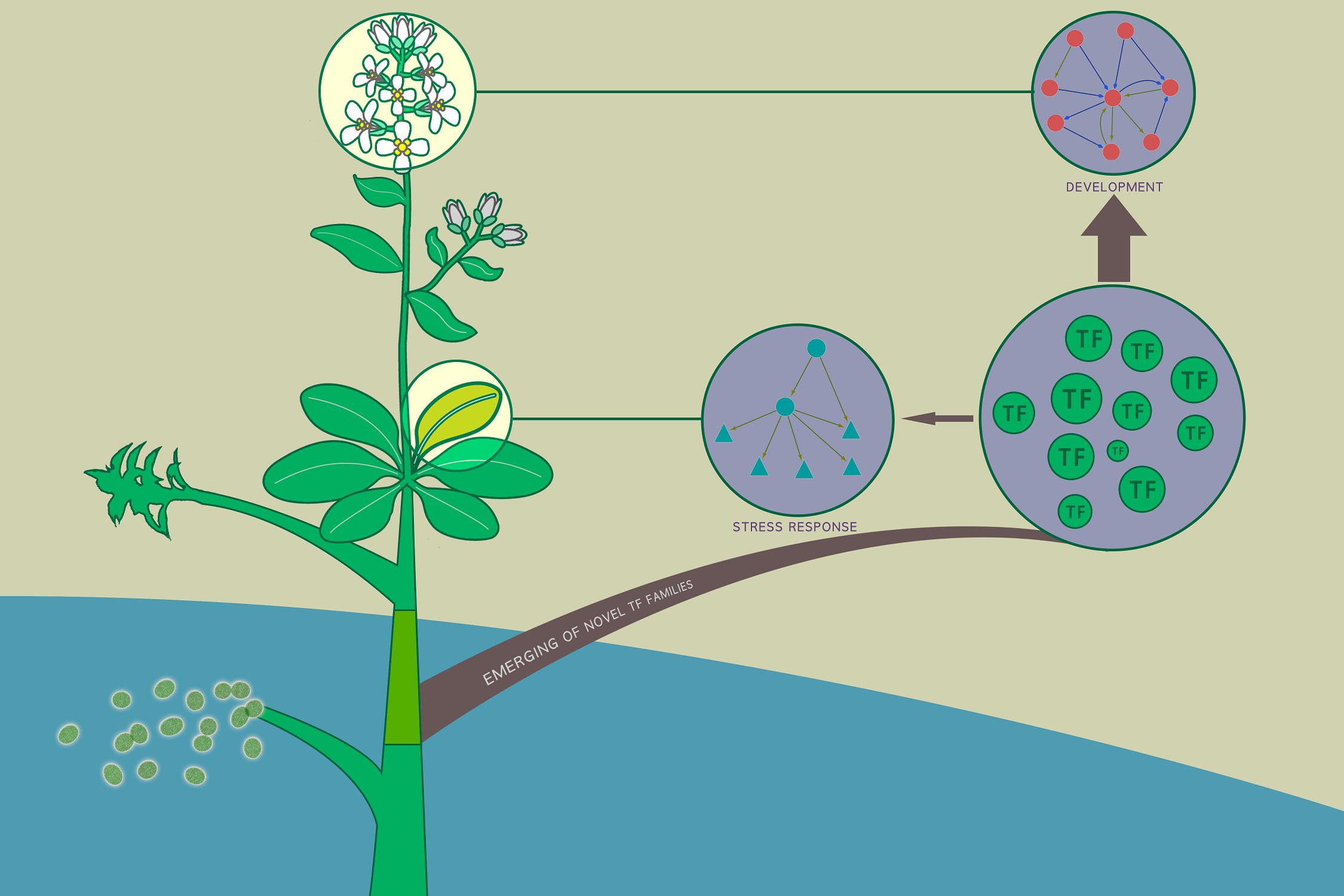
- Development vs stress-response: we reveal the architectural heterogeneity of developmental and stress-response sub-networks.
- Unicellular vs multicellular: we identify three types of novel network motifs that are absent from unicellular organisms, which are essential for multicellular development.
- Plants vs animals: we identify a novel type of network motif that is not enriched in metazoan transcriptional regulatory networks, which might play roles in the continuous organ regeneration of plants during postembryonic development.
- Ancient vs novel: we find that TFs of novel families present higher binding specificity and are preferentially wired into developmental processes and these novel network motifs.
- Property vs position: we unveil the underlying connection between the binding specificity of TFs and their wiring preference in networks, which is further validated in human, yeast, and E. coli, explaining the wiring preference of novel-family TFs.
- These findings provide insights into the fate determination of novel TFs and their contribution to the evolution of transcriptional regulatory networks.
See the corresponding manuscript [Full Text (PDF), Supplementary Materials]for more details.
Transcriptional regulatory mechanisms of multiple processes in
A. thaliana
, one of the best-studied model plants, have been reported in a large amount of scientific literature, providing an opportunity to build up a high-quality
Arabidopsis
transcriptional regulatory network. Here, we used two data sources,
PubMed Abstracts and
ResNet Plant 3.0 (A commercial knowledgebase of Pathway Studio mined from both PubMed Abstract and full-texts of plant-specific journal, now called
Pathway Studio Plant)to collect verified transcriptional regulatory interactions in
A. thaliana
. We retrieved transcription factor(TF)-associated interactions from ResNet Plant 3.0 as well as mined them from PubMed abstracts by MedScan. After manual curation each interaction by reviewing the original texts, we constructed an
Arabidopsis
transcriptional regulatory map (ATRM) that covers
388
TFs from
47
families, with directly supporting evidences from
974
references. This map mainly involves regulations in the developmental and stress response processes, providing a global landscape of transcriptional regulation in a plant species and also a valuable resource to study the architecture and evolution of plant transcriptional regulatory networks.
Through this website:
- ATRM can be visualized in kinds of styles and layouts.
- By double-clicking on nodes and edges, the information for corresponding genes and regulations are shown in the side panel, respectively. For genes, cross-links to PlantTFDB and TAIR for TFs and non-TFs are provided respectively. For regulations, cross-links to PubMed literature where regulations came are provided.
- Network data can be saved as different formats for further analysis.
How to Cite: